Salmonella spp.
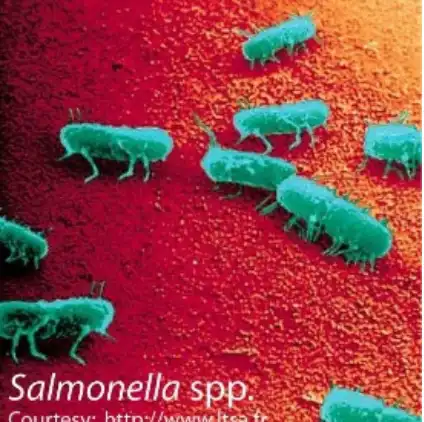
Scanning electron micrograph of Salmonella spp. The flagellae are clearly visible.
Samples:
Notes: Send all samples at room temperature, preferably preserved in sample buffer (PDF)
Interpretation of PCR Results:
Salmonella
Salmonella are motile, gram negative, aerobic, rod-shaped bacilli of the bacterial family Enterobacteriaceae. The genus Salmonella is obligately pathogenic and is divided into five subgenera that include approximately 2000 serovars. Salmonella can infect a wide variety of mammals, birds, reptiles, and other animals (Wigley, 2004).
Clinical Signs
Clinical signs for salmonellosis vary according to the host immune status, the number of infecting Salmonella, and the concomitant diseases (Wigley, 2004). Most cases of salmonellosis in animals are asymptotic, but infected animals may exhibit fever, poor appetite, diarrhea, vomiting, and abdominal pain. Generally, salmonellosis infection syndromes can be divided into persistence of an asymptomatic carrier state, gastroenteritis, bacteremia, endotoxemia, and organ localization of the infection.
Standard Diagnostic Methods
The diagnosis of salmonellosis is suspected in animals with acute or chronic gastrointestinal illness. However, this is difficult because the clinical and pathological findings of salmonellosis may be indistinguishable from those of other bacterial and viral diseases. Traditional diagnostic methods for salmonellosis include bacterial isolation, immunologic identification, serologic testing, and cytologic examination. While Salmonella culture remains the gold standard with some limitations, it may take up to five days to obtain final results (Uyttendaele et al., 2003).
Our Method
The Molecular Diagnostics Laboratory at Auburn University has developed a quantitative PCR technology targeting the highly conserved ttrC locus that is present in all Salmonella species and encodes the gene responsible for tetrathionate respiration of Salmonella. This test quickly detects both symptomatic and asymptomatic animals, and confirms the presence of as few as single Salmonella organisms in the PCR nucleic acid input. Our method detects all Salmonella strains with maximum sensitivity and highest specificity. In a study in leatherback turtle breeding grounds in the Caribbean, our method has helped to investigate Salmonella enterica infections (Dutton et al., 2013)
